Features
- ANAI Preprocessing
- ANAI AutoML
- Explainable ANAI
- Algorithms
- Anomaly Detection
- Ingesting data using inbuilt connectors
ANAI preprocessing pipeline
Initialization
import anai
from anai.preprocessing import Preprocessor
df = anai.load("data/bodyPerformance.csv", df_kwargs={"header": None})
prep = Preprocessor(dataset=df, target="class", except_columns=['weight_kg'])
Data Loading
Load data from a file
df = anai.load("data/bodyPerformance.csv", df_kwargs={"header": None}, legacy=False)
Returns a pandas dataframe
Available Preprocessing Methods
Data Summary
Gives a summary of the data.
summary = prep.summary()
#Returns a DataFrame
Column Statistics
Gives a column wise statistics of the dataset.
column_stats = prep.column_summary()
Returns a DataFrame
Imputing Missing Values
Imputes the missing values using the statistical methods.
df1 = prep.impute()
Returns a imputed DataFrame
Encoding Categorical Variables
Encodes the categorical variables.
df = prep.encode(split = False)
Returns a encoded DataFrame if split is False else returns a tuple of encoded Features and encoded Labels
Scaling Variables
Scales the variables using StandardScaler.
df = prep.scale(columns = [<List of Columns>], method = 'standard')
Available methods are 'standardize' and 'normal'
Returns a scaled DataFrame
Skewness Correction
Normalizes the skewness of the data.
df = prep.skewcorrect()
Returns a BoxCox Normalised DataFrame.
Prepare
Prepares the data for modelling.
X_train, X_val, y_train, y_val, scaler = prep.prepare(features, labels, test_size, random_state, smote, k_neighbors)
Arguments:
- features: pd.DataFrame or np.array
features to be used for training
- labels: pd.Series or np.array
labels to be used for training
- test_size: Size of the test set
- random_state: Random state for splitting the data
- smote: Boolean to use SMOTE or not
- k_neighbors: Number of neighbors to use for SMOTE
Returns:
- X_train: Training Features
- X_val: Validation Features
- y_train: Training Labels
- y_val: Validation Labels
- sc: Scaler Object
AutoML Pipeline
Initialization
import anai
ai = anai.run(filepath="data/iris.csv", target="class", predictor="lr")
Arguments
df : Pandas DataFrame
DataFrame to be used for modelling.
filepath : str
Filepath of the dataframe to be loaded.
df_kwargs : dict
Keyword arguments for the dataframe loading function. Only used if filepath is not None.
target : str
Target Column Name
except_columns : list, optional
List of Columns to be excluded from the dataset
predictor : list
Predicting models to be used
params : dict
dictionary containing parameters for model.
Not available when using predictor = all or multiple predictors.
test_size: float or int, default=.2
If float, should be between 0.0 and 1.0 and represent
the proportion of the dataset to include in
the test split.
If int, represents the absolute number of test samples.
cv_folds : int
No. of cross validation folds. Default = 10
pca : bool
if True will apply PCA on Train and Validation set. Default = False
lda : str
if True will apply LDA on Train and Validation set. Default = False
pca_kernel : str
Kernel to be use in PCA. Default = 'linear'
n_components_lda : int
No. of components for LDA. Default = 1
n_components_pca : int
No. of components for PCA. Default = 2
smote : Bool,
Whether to apply SMOTE. Default = True
k_neighbors : int
No. of neighbors for SMOTE. Default = 1
verbose : boolean
Verbosity of models. Default = False
exclude_models : list
List of models to be excluded when using predictor = 'all' . Default = []
path : list
List containing path to saved model and scaler. Default = None
Example: [model.pkl, scaler.pkl]
random_state : int
Random random_state for reproducibility. Default = 42
tune : boolean
when True Applies Optuna to find best parameters for model
Default is False
optuna_sampler : Function
Sampler to be used in optuna. Default = TPESampler()
optuna_direction : str
Direction of optimization. Default = 'maximize'
Available Directions:
maximize : Maximize
minimize : Minimize
optuna_n_trials : int
No. of trials for optuna. Default = 100
metric : str,
metric to be used for model evaluation. Default = 'r2' for regressor and 'accuracy' for classifier
suppress_task_detection: Bool
Whether to suppress automatic task detection. Default = False
task : str
Task to be used for model evaluation. Default = None
Only applicable when suppress_task_detection = True
Available Tasks:
classification : Classification
regression : Regression
Return
ai : regression or classification object
Returns a regression or classification object
Available Methods
Result
Gives the result of the model.
result = ai.result()
Returns a dataframe of the summary
Predict
Predicts the target column for the given data.
pred = ai.predict(data)
returns the predictions
Save Model
Saves the model to the given path.
path_model, path_scaler = ai.save(path = [path_model.pkl, path_scaler.pkl])
Returns the path to the model and scaler
Explain
Explains the model predictions.
exp = ai.explain(method = 'shap')
Avaliable Methods:
shap : SHAP
perm : Permutation
Examples
Load Model
Loads the model from the given path.
ai = anai.run(path = [path_model.pkl, path_scaler.pkl])
Hyperparameter Tuning
Use Tune=True to apply Optuna to find best parameters for model.
ai = anai.run(filepath="data/iris.csv", target="class", predictor="lr", tune = True)
All Models
Use predictor = 'all' to use all the models.
ai = anai.run(filepath="data/iris.csv", target="class", predictor="all")
Multiple Models
Pass a list of models to use to predictor.
ai = anai.run(filepath="data/iris.csv", target="class", predictor=['lr', 'rf'])
Principal Component Analysis
Use pca = True to use PCA on Train and Validation set.
ai = anai.run(filepath="data/iris.csv", target="class", predictor="lr", pca = True)
Linear Discriminant Analysis
Use lda = True to use LDA on Train and Validation set.
ai = anai.run(filepath="data/iris.csv", target="class", predictor="lr", lda = True)
Explainable ANAI
ANAI model predictions can be explained using SHAP or Permutation.
Initialize
import anai
ai = anai.run(filepath="data/iris.csv", target="class", predictor="lr")
Explain
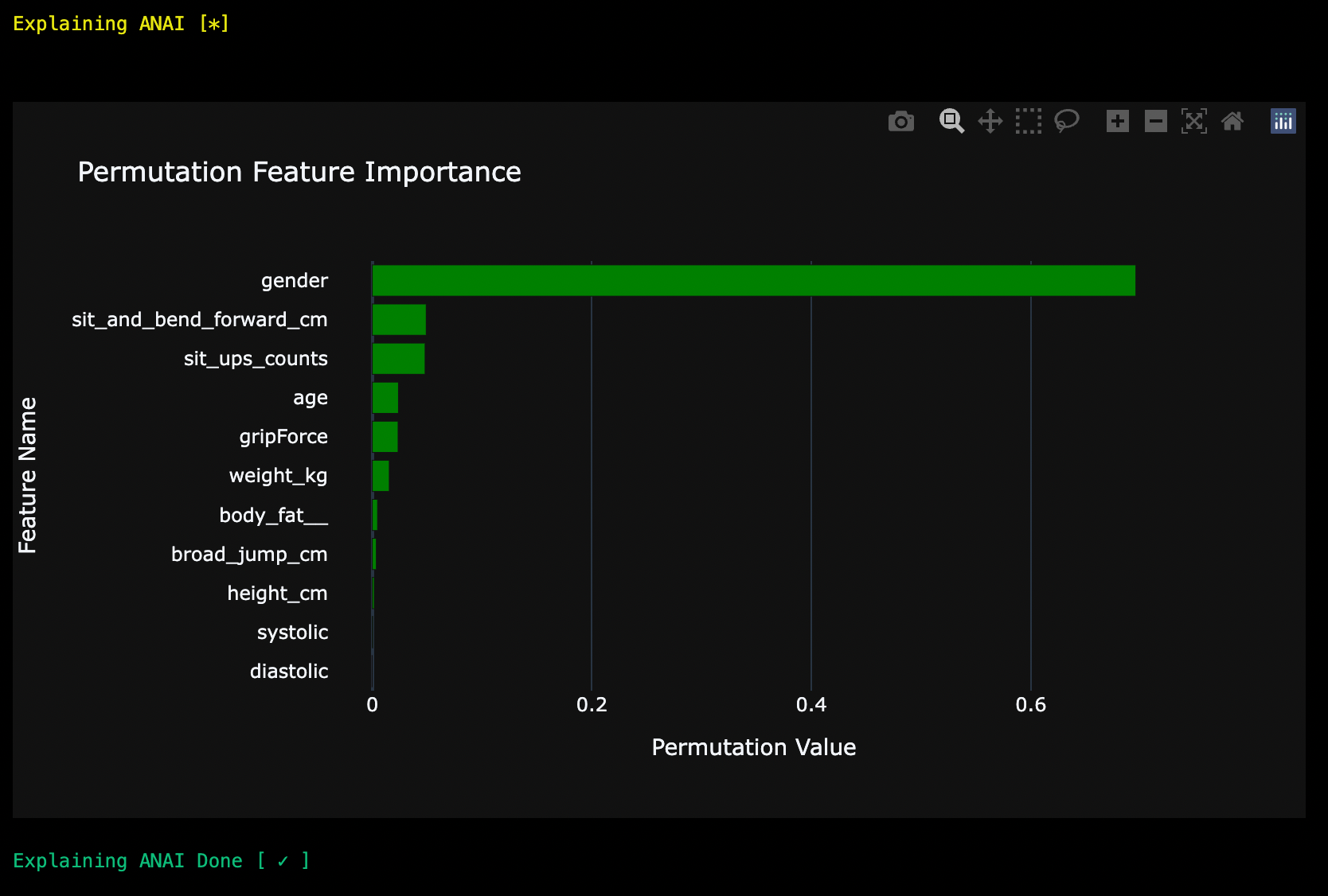
1) Permutation:
ai.explain(method = 'perm')
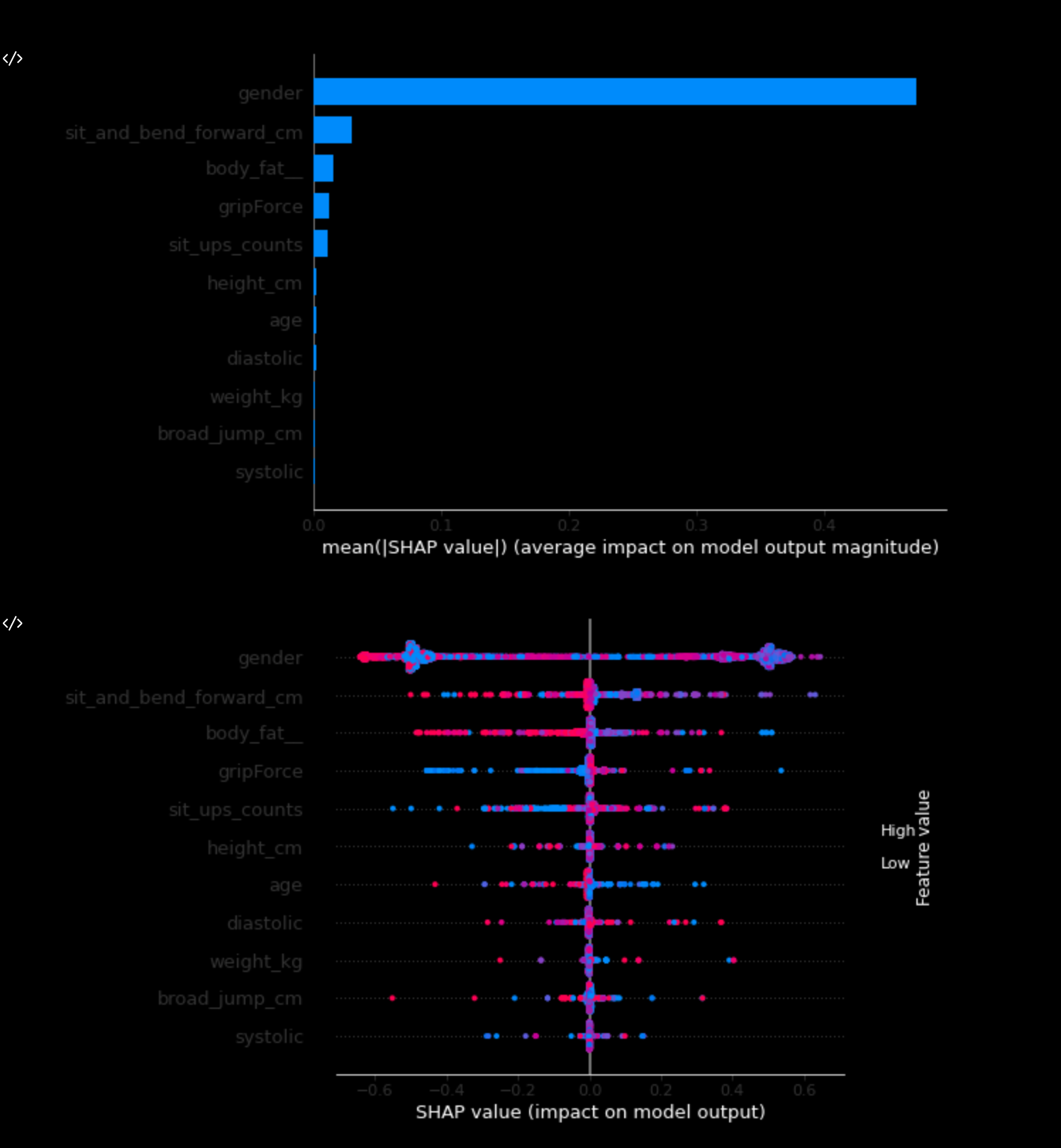
2) SHAP:
ai.explain(method = 'shap')
Surrogate Explainer
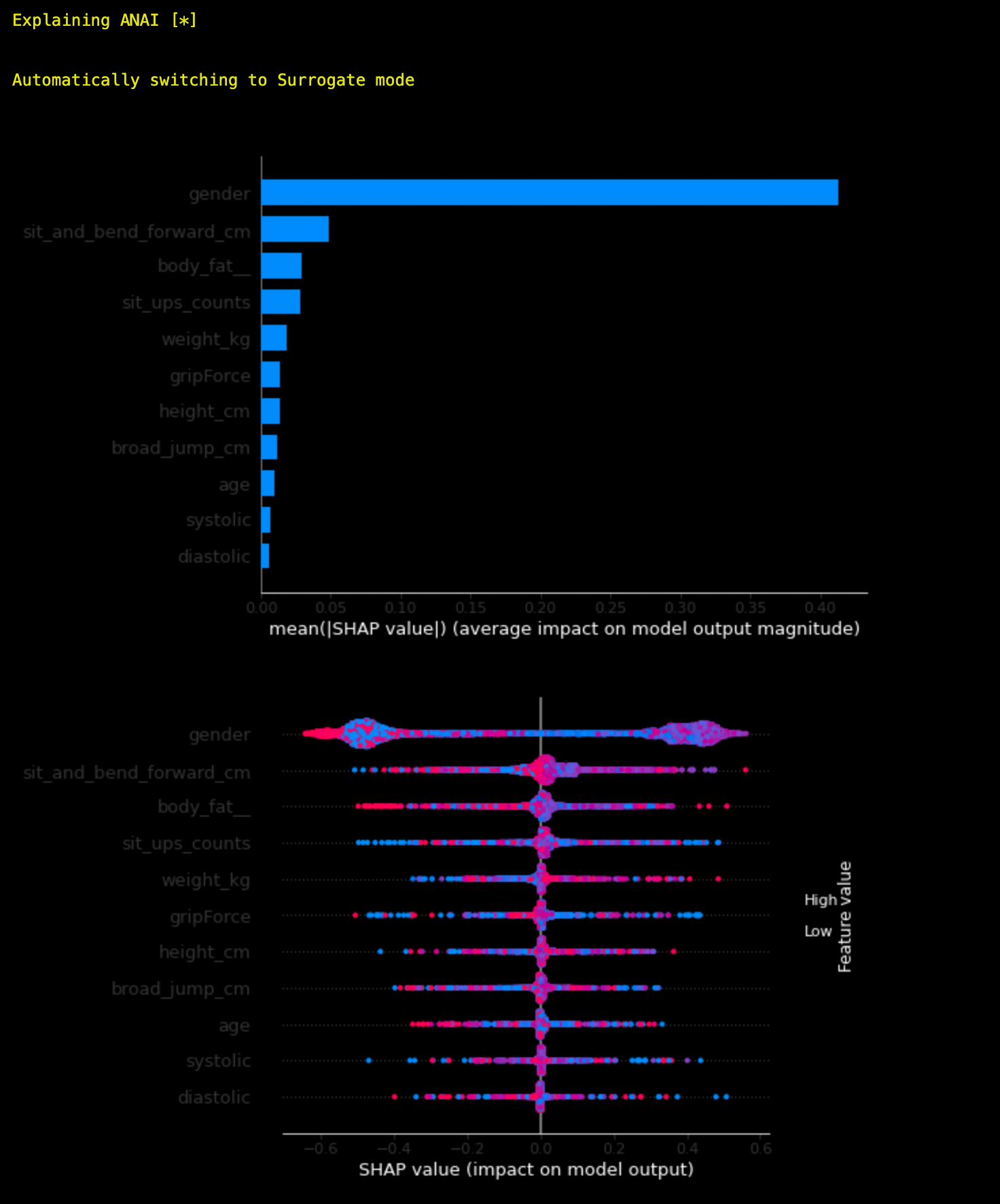
ANAI uses SHAP tree explainer to explain model predictions. So if the explainer fails to explain certain model it will switch to Surrogate Mode and use Decision Tree Surrogate Model to explain the original trained model. Surrogate mode is available for SHAP and Permutation exaplainers.
ai = anai.run(filepath="data/iris.csv", target="class", predictor="knn")
ai.explain(method = 'shap')
As you can see, SHAP explainer failed to explain the model. So it switched to Surrogate Mode and used Decision Tree Surrogate Model to explain the original trained model.
Available Algorithms
Regression
Available Models for Regression
- "lin": "Linear Regression"
- "sgd": "Stochastic Gradient Descent Regressor"
- "krr": "Kernel Ridge Regression"
- "elas": "Elastic Net Regression"
- "br": "Bayesian Ridge Regression"
- "svr": "Support Vector Regressor"
- "knn": "K-Nearest Neighbors"
- "dt": "Decision Trees Regressor"
- "rfr": "Random Forest Regressor"
- "gbr": "Gradient Boosted Regressor"
- "ada": "AdaBoostRegressor"
- "bag": "Bagging Regressor"
- "ext": "Extra Trees Regressor"
- "lgbm": "LightGBM Regressor"
- "xgb": "XGBoost Regressor"
- "cat": "Catboost Regressor"
- "ann": "Multi-Layer Perceptron Regressor"
- "poisson": "Poisson Regressor"
- "huber": "Huber Regressor"
- "gamma": "Gamma Regressor"
- "ridge": "Ridge CV Regressor"
- "encv": "ElasticNetCV Regressor"
- "lcv": "LassoCV Regressor"
- "llic": "LassoLarsIC Regressor"
- "llcv": "LassoLarsCV Regressor"
- "ransac": "RANSACRegressor",
- "ompcv": "OrthogonalMatchingPursuitCV",
- "gpr": "GaussianProcessRegressor",
- "omp": "OrthogonalMatchingPursuit",
- "llars": "LassoLars",
- "iso": "IsotonicRegression",
- "rnr": "Radius Neighbors Regressor Regressors",
- "qr": "Quantile Regression Regressors",
- "theil": "TheilSenRegressor Regressors",
- "all": "All Regressors"
Classification
Available Models for Classification
- "lr": "Logistic Regression"
- "sgd": "Stochastic Gradient Descent"
- "perc": "Perceptron"
- "pass": "Passive Aggressive Classifier"
- "ridg": "Ridge Classifier"
- "svm": "Support Vector Machine"
- "knn": "K-Nearest Neighbors"
- "dt": "Decision Trees"
- "nb": "Naive Bayes"
- "rfc": "Random Forest Classifier"
- "gbc": "Gradient Boosting Classifier"
- "ada": "AdaBoost Classifier"
- "bag": "Bagging Classifier"
- "ext": "Extra Trees Classifier"
- "lgbm": "LightGBM Classifier"
- "cat": "CatBoost Classifier"
- "xgb": "XGBoost Classifier"
- "ann": "Multi Layer Perceptron Classifier"
- "poisson": "Poisson Classifier"
- "huber": "Huber Classifiers"
- "ridge_cv": "RidgeCV Classifier"
- "encv": "ElasticNet CV Classifier"
- "lcv": "LassoCV Classifier"
- "llic": "LassoLarsIC Classifier"
- "llcv": "LassoLarsCV Classifier"
- "ransac": "RANSACClassifiers",
- "ompcv": "OrthogonalMatchingPursuitCV Classifier",
- "omp": "OrthogonalMatchingPursuit Classifier",
- "iso": "IsotonicRegression Classifier",
- "rad": "RadiusNeighbors Classifier",
- "quantile": "QuantileRegression Classifier",
- "theil": "TheilSenRegressor Classifier",
- "lars": "Lars Classifeir",
- "lcv": "LarsCV Classifier",
- "tweedie": "TweedieClassifiers",
- "all": "All Classifiers"
Model Explanation
Available Explanation Methods
- 'perm': Permutation Importance
- 'shap': Shapley Importance
Anomaly Detection
ANAI uses PYOD for anomaly detection.
Available Models for Anomaly Detection
- "IForest": "Isolation Forest"
- "CBLOF": "Cluster-based Local Outlier Factor"
Missing Data handling
ANAI supports statistical imputing
Available Methods for Missing Data Handling
- "mean": "Mean Imputation"
- "median": "Median Imputation"
- "mode": "Mode Imputation"
- "drop": "Drop Missing Data"
Categorical Encoding
ANAI uses catboost encoder for categorical feature encoding and Label Encoding for categorical target
Scaling
ANAI supports both standard and normalization scaling.
Available Scaling Methods
- "standardize": "Standard Scaling"
- "normalize": "Normalization Scaling"
Anomaly Detection
Usage
import anai
from anai.unsupervised import anomaly_detector
df = anai.load(filepath='data/iris.csv')
(anomaly_combined,
data_with_outliers,
data_with_Inliers,
anomaly_summary) = anomaly_detector(dataset = df, target = 'class', model = ['IForest', 'CBLOF'])
Returns
anomaly_combined : DataFrame
df modified with anomaly scores and anomaly
data_with_outliers : DataFrame
Only outliers
data_with_Inliers : DataFrame
Only Inliers
anomaly_summary : DataFrame
Summary of the anomaly detection
Ingesting data using the inbuilt connectors
Supported Sources
1) MySQL
2) S3
3) BigQuery
Examples
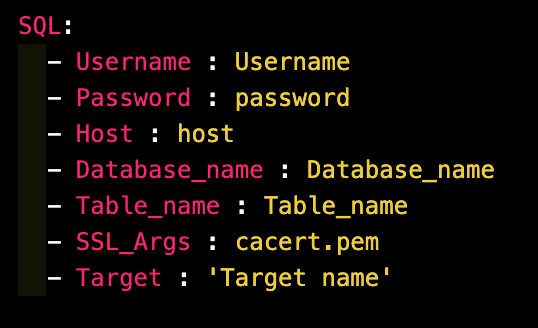
Example Config File for MySQL
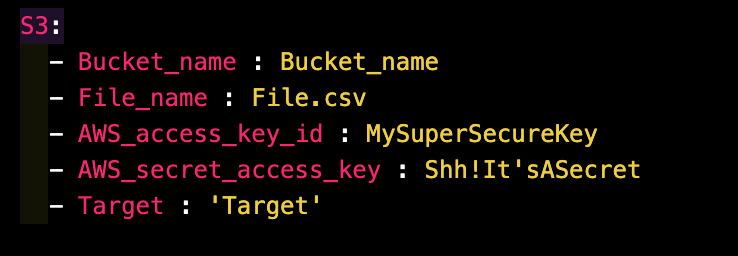
Example Config File for S3
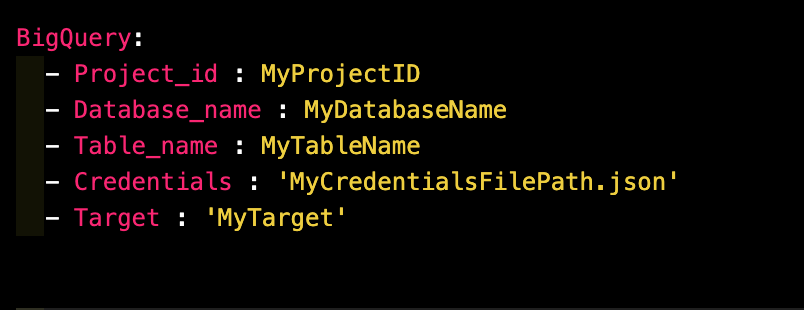
Example Config File for BigQuery
Run ANAI
ANAI will automatically detect the source of the data and run the appropriate connector using the config file. When Config = True. ANAI will automatically find the anai_config.yaml file in the current working directory.
import anai
ai = anai.run(config=True, predictor=['rfr'])